The GPU Advances: ATI's Stream Processing & Folding@Home
by Ryan Smith on September 30, 2006 8:00 PM EST- Posted in
- GPUs
Folding@Home
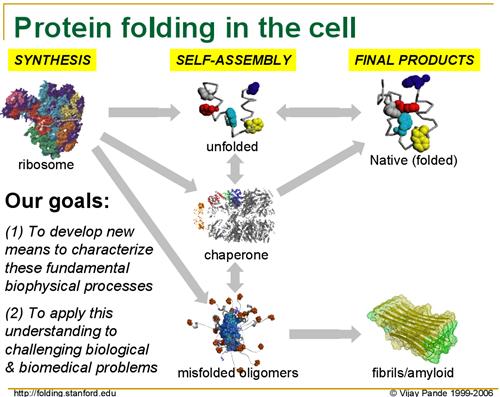
For several years now, Dr. Vijay Pande of Stanford has been leading the Folding@Home project in order to research protein folding. Without diving unnecessarily into the biology of his research, as proteins are produced from their basic building blocks - amino acids - they must go through a folding process to achieve the right shape to perform their intended function. However, for numerous reasons protein folding can go wrong, and when it does it can cause various diseases as malformed proteins wreck havoc in the body.
Although Folding@Home's research involves multiple diseases, the primary disease they are focusing on at this point is Alzheimer's Disease, a brain-wasting condition affecting primarily older people where they slowly lose the ability to remember things and think clearly, eventually leading to death. As Alzheimer's is caused by malformed proteins impairing normal brain functions, understanding how exactly Alzheimer's occurs - and more importantly how to prevent and cure it - requires a better understanding on how proteins fold, why they fold incorrectly, and why malformed proteins cause even more proteins to fold incorrectly.
The biggest hurdle in this line of research is that it's very computing intensive: a single calculation can take 1 million days (that's over 2700 years) on a fast CPU. Coupled with this is the need to run multiple calculations in order to simulate the entire folding process, which can take upwards of several seconds. Even splitting this load among processors in a supercomputer, the process is still too computing intensive to complete in any reasonable amount of time; a processor will simulate 1 nanosecond of folding per day, and even if all grant money given out by the United States government was put towards buying supercomputers, it wouldn't even come close to being enough.
This is where the "@Home" portion of Folding@Home comes in. Needing even more computing power than they could hope to buy, the Folding@Home research team decided to try to spread processing to computers all throughout the world, in a process called distributed computing. Their hopes were that average computer users would be willing to donate spare/unused processor cycles to the Folding@Home project by running the Folding@Home client, which would grab small pieces of data from their central servers and return it upon completion.
The call for help was successful, as computer owners were more than willing to donate computer cycles to help with this research, and hopefully help in coming up with a way to cure diseases like Alzheimer's. Entire teams formed in a race to see who could get more processing done, including our own Team AnandTech, and the combined power of over two-hundred thousand CPUs resulted in the Folding@Home project netting over 200 Teraflops (one trillion Floating-point Operations Per Second) of sustained performance.
While this was a good enough start to do research, it was still ultimately falling short of the kind of power the Folding@Home research group needed to do the kind of long-runs they needed along side short-run research that the Folding@Home community could do. Additionally, as processors have recently hit a cap in terms of total speed in megahertz, AMD and Intel have been moving to multiple-core designs, which introduce scaling problems for the Folding@Home design and is not as effective as increasing clockspeeds.
Since CPUs were not growing at speeds satisfactory for the Folding@Home research group, and they were still well short of their goal in processing power, the focus has since returned to stream processors, and in turn GPUs. As we mentioned previously, the massive floating-point power of a GPU is well geared towards doing research work, and in the case of Folding@Home, they excel in exactly the kind of processing the project requires. To get more computing power, Folding@Home has now turned towards utilizing the power of the GPU.
For several years now, Dr. Vijay Pande of Stanford has been leading the Folding@Home project in order to research protein folding. Without diving unnecessarily into the biology of his research, as proteins are produced from their basic building blocks - amino acids - they must go through a folding process to achieve the right shape to perform their intended function. However, for numerous reasons protein folding can go wrong, and when it does it can cause various diseases as malformed proteins wreck havoc in the body.
 |
| Click to enlarge |
Although Folding@Home's research involves multiple diseases, the primary disease they are focusing on at this point is Alzheimer's Disease, a brain-wasting condition affecting primarily older people where they slowly lose the ability to remember things and think clearly, eventually leading to death. As Alzheimer's is caused by malformed proteins impairing normal brain functions, understanding how exactly Alzheimer's occurs - and more importantly how to prevent and cure it - requires a better understanding on how proteins fold, why they fold incorrectly, and why malformed proteins cause even more proteins to fold incorrectly.
The biggest hurdle in this line of research is that it's very computing intensive: a single calculation can take 1 million days (that's over 2700 years) on a fast CPU. Coupled with this is the need to run multiple calculations in order to simulate the entire folding process, which can take upwards of several seconds. Even splitting this load among processors in a supercomputer, the process is still too computing intensive to complete in any reasonable amount of time; a processor will simulate 1 nanosecond of folding per day, and even if all grant money given out by the United States government was put towards buying supercomputers, it wouldn't even come close to being enough.
This is where the "@Home" portion of Folding@Home comes in. Needing even more computing power than they could hope to buy, the Folding@Home research team decided to try to spread processing to computers all throughout the world, in a process called distributed computing. Their hopes were that average computer users would be willing to donate spare/unused processor cycles to the Folding@Home project by running the Folding@Home client, which would grab small pieces of data from their central servers and return it upon completion.
The call for help was successful, as computer owners were more than willing to donate computer cycles to help with this research, and hopefully help in coming up with a way to cure diseases like Alzheimer's. Entire teams formed in a race to see who could get more processing done, including our own Team AnandTech, and the combined power of over two-hundred thousand CPUs resulted in the Folding@Home project netting over 200 Teraflops (one trillion Floating-point Operations Per Second) of sustained performance.
While this was a good enough start to do research, it was still ultimately falling short of the kind of power the Folding@Home research group needed to do the kind of long-runs they needed along side short-run research that the Folding@Home community could do. Additionally, as processors have recently hit a cap in terms of total speed in megahertz, AMD and Intel have been moving to multiple-core designs, which introduce scaling problems for the Folding@Home design and is not as effective as increasing clockspeeds.
Since CPUs were not growing at speeds satisfactory for the Folding@Home research group, and they were still well short of their goal in processing power, the focus has since returned to stream processors, and in turn GPUs. As we mentioned previously, the massive floating-point power of a GPU is well geared towards doing research work, and in the case of Folding@Home, they excel in exactly the kind of processing the project requires. To get more computing power, Folding@Home has now turned towards utilizing the power of the GPU.










43 Comments
View All Comments
photoguy99 - Sunday, October 1, 2006 - link
Basic research is like that - it may takes a lot of years to benefit from it.Look at Einstein, his work was fundamental research but the benefits are still being realized 100 years later.
So even if they have major breakthroughs they may be at such a foundational level that the actual cure for Alz. comes 25 years later.
Nature of the beast.
JarredWalton - Sunday, October 1, 2006 - link
http://folding.stanford.edu/results.html">Published ResultsCurrent research includes:
http://folding.stanford.edu/FAQ-diseases.html#AD">Alzheimer's, Cancer, Huntington's Disease, Osteogenesis Imperfecta, Parkinson's Disease, Ribosome and antibiotics.
And of course, there's always http://folding.stanford.edu/faq.html">the Folding@Home FAQ.
Do they know in advance that all of the issues are related to protein folding? No, but I'd assume they have good cause to suspect it. The problem is that it takes time; breakthrough results might not materialize soon, next year, or even for 5-10 year. Should research halt just because the task is difficult? Personally, I think FAH has a far greater chance of impacting the world during my lifetime than SETI@Home.
Cheers!
Baked - Sunday, October 1, 2006 - link
I wonder if a X1600 card will work. I've tried both the graphics and command line version of F@H on my new system but both had problem connecting to F@H server. Hopefully this new F@H version will work.JarredWalton - Sunday, October 1, 2006 - link
Next step is to extend to X1800 and probably from there to X1600. Beyond that, the G70 chips are probably the next up would be my guess.smitty3268 - Sunday, October 1, 2006 - link
I assume the new client uses the cpu + gpu, and not just the gpu? Also, it would be nice to have some sort of explanation for the poor nvidia performance in the next article. Is it just their architecture, or has Folding@Home been getting assistance from ATI and not NVidia?This doesn't make much sense:
The Folding@Home design is quite obviously a massivly parallel design, as shown by the fact that hundreds of thousands of computers are all working on the same problem. Therefore, doubling the amount of cores would double the amount of work being done and this seems to be happening faster than the old incremental speed bumps.
Otherwise, it was a good article.
z3R0C00L - Monday, October 2, 2006 - link
It's Simple.. F@H uses Dynamic Branching Calculations. nVIDIA GPU's are technologically inferior to ATi VPU's when it comes to shading performance and branching performance.As such.. nVIDIA's highly mighty GeForce 7950GX2 would perform much like an ATi Radeon x1600XT. In other words.. too slow.
tygrus - Monday, October 9, 2006 - link
The Nvidia FP hardware is fast enough but the overall design doesn't fit well with the software(task) design of F@H. For other tasks the Nvidia GPU's may be very fast. The next Nvidia GPU & API will hopefully be better.The CPU handles the data transformation and setup before sending to GPU for the accelerated portion. Then the CPU overseas the return of data from the GPU. The CPU also looks after the log, text console, disk read/write, internet upload&download, and other system ovreheads.
More information is available from ???
i just found a really great article which covers the public release: http://techreport.com/onearticle.x/10907">http://techreport.com/onearticle.x/10907
quote:
--------------------------------------------------------------------------------
Talk w/Vijay Pande
ATI is currently 8X faster than Nvidia. Nvidia has our code, running it internally, hope we can close the gap. But even 4X difference is large, and ATI is getting faster all of the time.
Lot of work goes into qualifying GPUs internally so they can run.
Making apps like this run on a GPU requires a lot of development work. Currently, science is best served by using ATI chips. Nv may come in future.
---------
The CPU has to poll the GPU to find out if it finished a block and needs help (data from GPU->CPU etc). This takes a context switch and CPU time to wait for the reply (ns wait not fixed number of cycles). Any acutual work is done in the remaining time slice or more. The faster the GPU, the more it demands uses the CPU. Slow the CPU in half and you may be slowing GPU by upto half.
Ryan Smith - Sunday, October 1, 2006 - link
The new client "uses" the CPU like all applications do, but the core is GPU-based, so it won't be pushing the CPU like it does on the CPU-only client, I don't know to what level that means however.As for the Nvidia stuff, we only know what the Folding team tells us. They made it clear that the Nvidia cards do not show the massive gains that ATI's cards do when they try to implement their GPU code on Nvidia's cards. Folding@Home has been getting assistance from Nvidia, but they also made it clear that this is something they can do without help, so the problem is in the design of the G7x in executing their code.
As for the core stuff, this is something the Folding team explicitly brought up with us. The analogy they used is trying to bring together 2000 grad students to write a PhD thesis in 1 day, it doesn't scale like that. They can add cores to a certain point, but the returns are diminishing versus faster methods of processing. This is directly a problem for Folding@Home, which is why they are putting efforts in to stream processing, which can offer the gains they need.
smitty3268 - Sunday, October 1, 2006 - link
Does the G7x have as much support for 32bit floats as ATI does? It seems like I read somewhere that one of the two had moved to 32 bit exclusively while the other was still much faster at 16/24 bit fp. Could that be why they aren't seeing the same performance from NVidia?Clauzii - Monday, October 2, 2006 - link
Probably that, and the fact that the big ATI models contain 48 shaders - pretty beefes the calculations up!